Custom Taqman Assay Design Tool
Efficient qPCR relies on good assay design. Since the invention of PCR, many parameters have been identified as important for assay quality, such as estimates of oligo temperature characteristics, GC content and folding properties. Empirical evidence has been used to demonstrate the value of each parameter and there are now assay design algorithms that predict good assay designs with high confidence. The high number of parameters and the complexity of their relationships make using software tools a huge advantage, rather than attempting assay design manually. Fortunately, there are now tools available online that are free of charge and yet incorporate the most advanced design algorithms. We provide OligoArchitect™ Online, free of charge. OligoArchitect Online is powered by Beacon Designer™ (PREMIER Biosoft International). Beacon Designer is well-known in the qPCR community and is often considered to be the best design software available. OligoArchitect Online contains many features (including the capability to design oligos modified with LNA®) of Beacon Designer, although not all. In this protocol, the capabilities of OligoArchitect will be demonstrated to find SYBR® Green I, End-point-PCR, Dual-Labeled Probe, Multiplex and SNP assay designs.
We offer both online and consultative assay design services. The team that supports the consultative assay designs is experienced and has access to many state-of-the-art assay design tools, including full versions of Beacon Designer. Consultative designs may be required to analyze sequence alignments, to design for conserved regions, or to design for specific targets. The consultative design team also utilizes tools to find designs across exon junctions, though this can be replicated manually using the online tool (directions can be found on the web page). These are examples of capabilities within consultative designs that go beyond the capabilities of the online tool, though the online tool is upgraded at regular intervals.
The two alternatives thus complement each other. The consultative design service provides extensive capabilities and personal assistance. OligoArchitect Online provides an easy-touse solution under your direct control.
Login and Registration
The OligoArchitect Online design tool is available by clicking the "Online Design" button. The homepage has detailed information about design parameters and version capabilities. Registration is completed by adding a few contact details and choosing a password.
Once logged in, there are several options available to load the target sequence. First, select the type of detection chemistry from the tabs at the top of the page. Options include SYBR Green I, Dual-Labeled Probe, Molecular Beacon, LightCycler® Probe and Scorpion® Probe (Figure 1). After selecting the desired detection chemistry, either enter an accession number or copy-paste the target sequence itself. After entering an accession number, simply click "Search" to run a design or the "Load Sequence" button to populate the target sequence window with the desired target sequence (this is only necessary to add a SNP).

Figure 1. SYBR Green I Assay Design.
Human GAPDH will be used as the target for the SYBR Green I Assay demonstration. Enter the accession number of human GAPDH (NM_002046) and load the target sequence. Click the "Search" button to use the default set of parameters to obtain design results. The result is presented in two ways. First, the sense and anti-sense primers are indicated with blue and red fonts, respectively, in the target sequence. Second, the primers are listed below the target sequence window together with GC content, temperature characteristics, and other oligo properties (Figure 1). The property "Rating" is the estimate that is calculated by the design algorithm for the design quality. Ratings above 50 are considered "good" and normally pass the algorithm criteria for approval to use. Ratings above 75 are listed as "best" and, thus, give extra confidence in the design quality.
Clicking the "View All Results" button allows the user to consider other alternative designs (Figure 2). This may be useful if the user has specific preferences that are not covered by the design parameters. Selection of the design is done by toggling the check box on the left-hand side of the design parameters table. Once satisfied with the selection, results can be exported to a local Microsoft® Excel file by clicking "Export for Synthesis" or entered directly into the Sigma online ordering tool for immediate ordering by clicking "Place an Order for Synthesis".

Figure 2. Primer Selection Indicated by Red and Blue Fonts.

Figure 3. Alternative Primer Options.
End-point PCR Assay Design
Like SYBR Green I assay designs, end-point PCR assay designs are also based on two, usually non-modified, oligos that prime PCR amplification events. The difference is that amplification detection occurs by identification on a gel or by a fragment analysis instrument for end-point PCR. This means that the length of the amplification product may need to be larger than for SYBR Green I qPCR assays. To modify the amplicon length, rather than clicking on "Search" immediately (as described above), the "Primer Parameters" link is opened, then scroll down and adjust the parameter for "Amplicon Length" from the default setting to a range from 300 to 2,000 nucleotides long (Figure 4) before selecting "Search". In the example of the human GAPDH (NM_002046) design, the resulting assay has an amplicon of 352 nucleotides long (Figure 5).

Figure 4. Adjusting Design Parameters for End-point PCR Detection.
![]()
Figure 5. Longer Amplicon Indicated by Primers (Blue and Red) Suitable for End-point PCR.
Dual-Labeled Probe Design
Together with SYBR Green I assay designs, Dual-Labeled Probe assay designs are the most common. Dual-Labeled Probe assays have the potential to be more sensitive and specific than SYBR Green I assays, although the additional oligo and modifications usually make these assays more expensive.
To perform the design using OligoArchitect, switch to "Dual-Labeled Probe" detection chemistry among the tabs at the top of the page (Figure 6). Add the human GAPDH (NM_002046) target sequence as described in the SYBR Green I design example above and click "Search" to generate new assay results. Again, the sense and anti-sense primers are indicated with blue and red font, respectively, in the target sequence. Additionally, we now also have the probe oligo indicated with purple font in the target sequence (Figure 7).
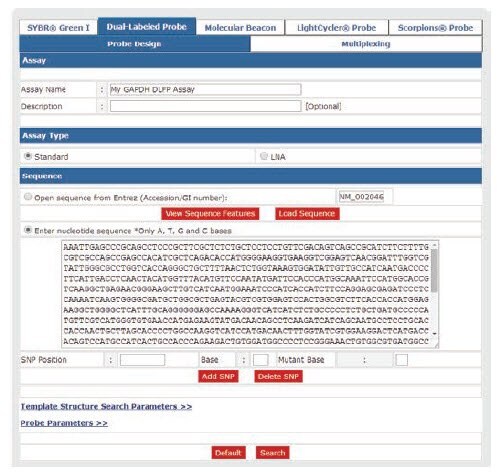
Figure 6. Adding Target Sequence for Dual-Labeled Probe Design.

Figure 7. Assay Design Showing Two Primers in Red and Blue and the Dual-Labeled Probe in Purple.
The potential to design multiplex assays is another advantage of Dual-Labeled Probe assays, when compared to SYBR Green I assays. Therefore, click "View All Results" and there is now also the option to have the design result "Save for Multiplexing" (Figure 8).

Figure 8. Selecting Dual-Labeled Probe Assays for Multiplex Analysis.
Multiplex Assay Design
After selecting "Save for Multiplexing" in the OligoArchitect Online tool, a window is presented that allows the assays that have been saved for multiplex analysis to be inspected. The "View Multiplex Oligos" leads to another window where the predicted risks of undesirable oligo cross reactions can be viewed. In the case of the GAPDH assay, there are three potential, undesirable oligo cross reactions predicted (Figure 9).

Figure 9. Predicting Oligo Interactions in Multiplex Reactions.
Having saved the GAPDH design for multiplex, click the "Back" button until returning to the window where another target sequence is added to the analysis. In this example, human HPRT1 (NM_000194) is loaded. Click "Search" to obtain the design results. Click "View All Results" and "Save for Multiplexing". Now there are saved assays for both GAPDH and HPRT1 and their multiplex compatibility can be evaluated. Select "View Multiplex Oligos" to reveal that, in this case, there are 14 potential, undesirable oligo cross reactions predicted (Figure 10).
The procedure described above was repeated to add human GUSB (NM_000181) to the multiplex assay. With three target genes in the multiplex, there are 35 undesirable oligo cross reactions predicted (Figures 11A, 11B, and 11C). In general, the risk of undesirable, oligo cross interactions increases exponentially with the number of oligos in the assay. Therefore, it is prudent to avoid higher orders of multiplex assay, if possible, considering other limitations of the assay.

Figure 10. Potential Oligo Interactions for Two Genes in Multiplex.

Figure 11A. Potential Oligo Interactions for Three Genes in Multiplex.

Figure 11B. Potential Oligo Interactions for Three Genes in Multiplex (cont'd).

Figure 11C. Potential Oligo Interactions for Three Genes in Multiplex (cont'd).
SNP Assay Design
Another common application for qPCR is SNP genotyping. OligoArchitect Online handles SNPs easily by allowing users to type in the position and mutant base below the target sequence window. For the presented example, the target sequence for human GUSB gene and a SNP site rs202210104 is used. The mutant position is 201, and the SNP is a T>C variation. Click the "Add SNP" button to activate the entered SNP. The activation of the selection is visually indicated by a green tick mark that appears on the right side of the SNP information (Figure 12).

Figure 12. Entering SNP Information to Target Sequence.
Click "Search" to proceed with the assay prediction routine. In this example, there is a highlighted message indicating that "No Probe found: 84 rejected (Gs > Cs: 84)" (Figure 13). This is a relatively common warning. The reason for this error message is that one of the characteristics of a good design is that the number of Gs should be less than the number of Cs for a good probe design (see PCR qPCR dPCR Assay Design). The solution for this concern is to encourage the design algorithm to find a suitable probe on the opposite strand of the target sequence. Click "Back" and then open the details of the "Probe Parameters >>". Scroll down and note that "Design Sense Probe" is activated under the section "Output Options". Change this to "Design Anti-sense Probe" by toggling the appropriate radio button (Figure 14). Click "Search" again to engage the design algorithm prediction routine. This time the search has yielded resulting designs with the probe targeting the antisense strand of the target sequence (Figure 15).

Figure 13. Error Message Indicating that No Suitable Probe Could be Designed (Gs > Cs).

Figure 14. Design Parameters Showing Selection of Sense and Anti-sense Probe Design.

Figure 15. Successful Anti-sense Probe Design.
Summary
In this protocol, the application of OligoArchitect Online has been demonstrated for the design of primers suitable for use with SYBR Green I and end-point-PCR assays; Dual-Labeled Probes for single as well as multiplex assays and assays targeting a specific SNP. More features are available within OligoArchitect Online that are beyond the scope of this protocol, including application of Molecular Beacons, LightCycler® Probes and Scorpions® Probes. For specific capabilities outside those available in OligoArchitect Online, consult the Sigma Custom Products design teams.
Oligo technical team
Custom Taqman Assay Design Tool
Source: https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/genomics/qpcr/oligoarchitect-assay-design
Posted by: havilandfert1948.blogspot.com

0 Response to "Custom Taqman Assay Design Tool"
Post a Comment